Weeknote 14/07/25
scAnt up and running
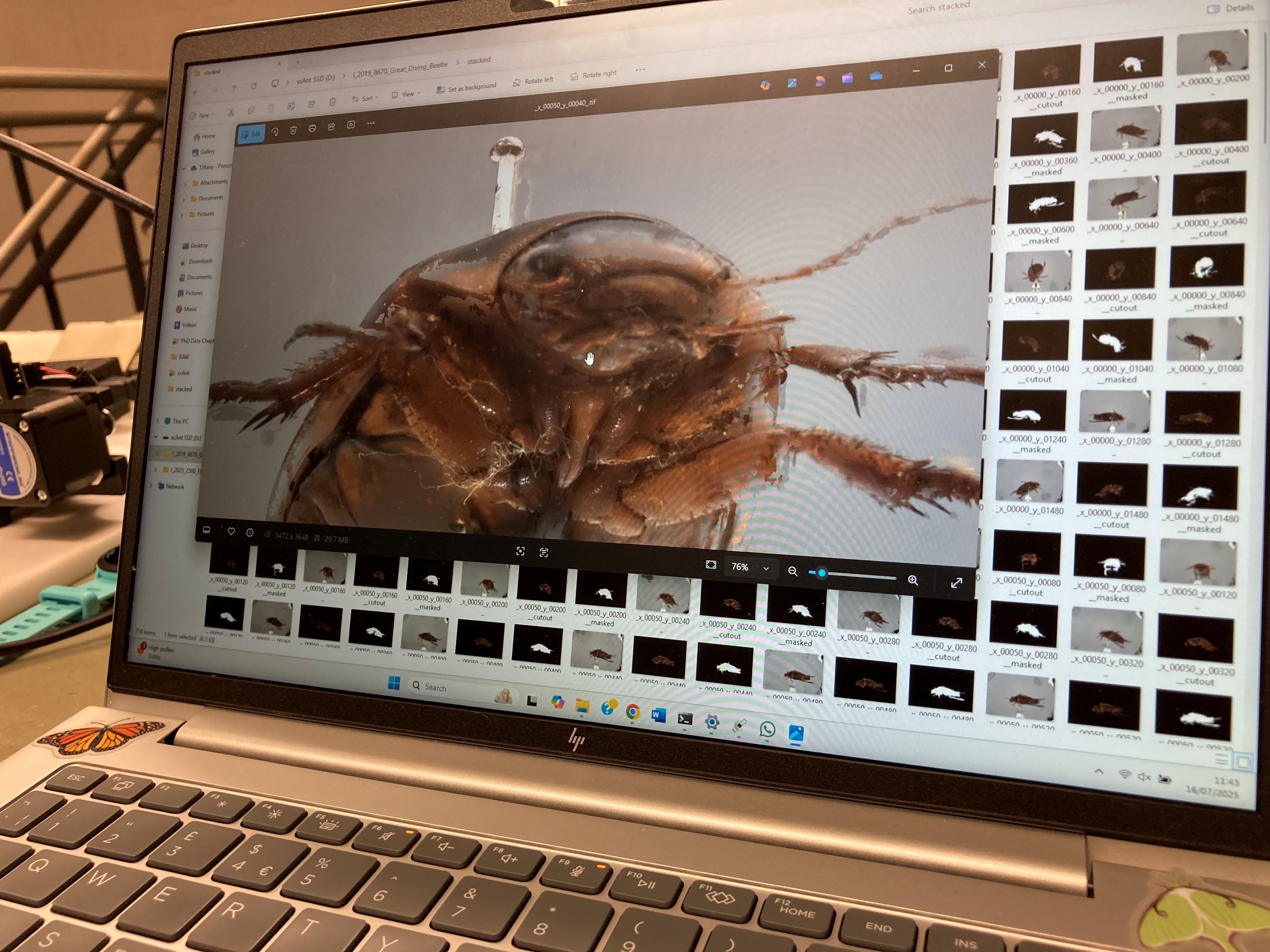
Since the scAnt has been set up this week, we have scanned four (currently scanning the fifth) specimens! We scanned an Elephant Hawk-moth, Great Diving Beetle, White-tailed Bumblebee and a Julodis viridipes beetle from South Africa. We are currently scanning a Large Blue, a species that went extinct in the UK in 1979 but has since been successfully reintroduced. On the Invertebrate Ecology gaming PC, Tiff currently has Meshroom running to create a 3D model via photogrammetry of the Great Diving Beetle. Alongside capturing these specimens with scAnt we have been scanning them on Polycam under limiting conditions like maximum number of photos with the aim of comparing the two methods.




Experimenting with React Native and iOS hacking for displaying images with cataloging information
Bea has been learning Swift and using XCode to create an iPhone app to display 2D imaging of specimens alongside their cataloging information like species and label data. This can be extended easily to display 3D models using RealityKit, which I will do next week.
Similarly, Arissa has started writing an app using React Native with Expo for a similar purpose, using Google Drive to store the photos taken and resulting 3D models, and display them on a scans page. So far, the structure and UI of the app is mostly done, and Google sign in is working on the web version (currently working on getting the mobile versions functional as well). There is a lot of work left to be done, especially with getting camera permissions and taking photos in the app, but I found an expo package that can be used for this, and it is the next task to be done once the Google Drive upload/display of files is working.
We based the colour scheme of the app off of Spanish moon moths:


Researching open source gaussian splatting variations
We’ve looked into a range of Gaussian Splatting variations including reading Differentiable Point-Based Radiance Fields for Efficient View Synthesis, 3D Gaussian Splatting for Real-Time Radiance Field Rendering, and GaussianObject: High-Quality 3D Object Reconstruction from Four Views with Gaussian Splatting. These discuss a range of improvements of processing and rendering speed and capture quality as compared to traditional Gaussian Splatting and reading them helped to gain an understanding of the general method behind Gaussian Splatting. We haven’t yet tried to run any of the code provided in the repos, since we have insufficient VRAM on our laptops. However, today Tiff logged into the gaming PC that the Invertebrate Ecology group can access for photogrammetry and similar applications, so we will look into running some trials on that.
Following on from our Thursday meeting with Anil and Sadiq, we looked into On Scaling Up 3D Gaussian Splatting Training, a new paper out of NYU on distributed Gaussian Splatting, and Brush, which is a 3D reconstruction engine using gaussian splatting. Brush is trained on posed image data (which can be done on mobile or in a browser as well), and can be used to display gaussian splats if they are in the specific .ply or .compressed.ply format (the splats can also be displayed on the web).
Insect corner
On Tuesday, we went to the DAB moth trapping again. There were so many moths this week! We saw a large range of macro- and micromoths including an Elephant Hawk-moth, Large, Broad-bordered and Lesser broad-bordered Yellow Underwings and Bird-Cherry Ermine moths. I was able to hold a Dark Arches moth and an Elephant Hawk-moth!



The moth traps use egg cartons as places for the moths to land and rest once they are in the moth trap:



Insect fact of the week: Only a handful of animals have colour vision at night, the Elephant Hawk-moth is one of them! The others are two more types of hawk-moths, the carpenter bee and a nocturnal gecko.


Thursday meetings
On Thursday we had a lab meeting with the Insect Ecology and Agroecology groups.
Tiff and Matt gave both groups a tour of stores with a focus on scAnt, which was scanning Julodis viridipes. After this, we chatted briefly with Jerry Liu, another UROP student in the Insect Ecology group about 3D reconstruction techniques as they relate to our respective projects.
Following this, we met with Tiff, Anil, and Sadiq to discuss our findings from the last two weeks and the direction going forward. This was useful in introducing us to On Scaling Up 3D Gaussian Splatting Training and Brush. We also discussed sending different sizes and resolutions of data to NYU for processing, with the aim of determining the number of images required and how high quality they had to be for certain morphological features to be identified and measured.